David Castells-Rufas, Santiago Marco-Sola, Juan Carlos Moure, Quim Aguado, Antonio Espinosa
Pre-alignment filters propose useful methods to reduce the computational requirements of genomic sequence mappers.
Most of the filters are based on estimating or actually computing the edit distance between the input reads and their candidate locations in the reference genome using a subset of the elements of the dynamic programming table used to compute the Levenshtein distance between the reads and the reference sequence.
Some FPGA implementations of pre-alignment filters use classic Hardware Description Language (HDL) toolchains, thus limiting their portability. Nowadays, C/C++ High Level Synthesis (HLS) is supported on most FPGA accelerators used by heterogeneous cloud providers. We have reimplemented and optimized several state-of-the-art pre-alignment filters using C/C++ based HLS to expand their portability to a wide range of systems supporting the OpenCL runtime.
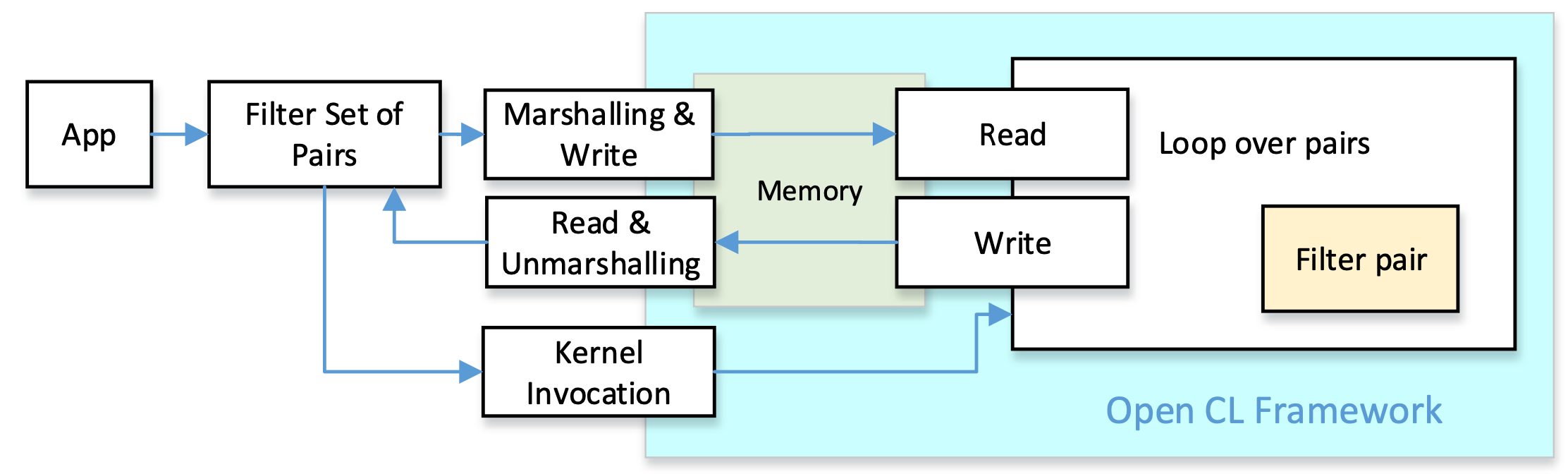
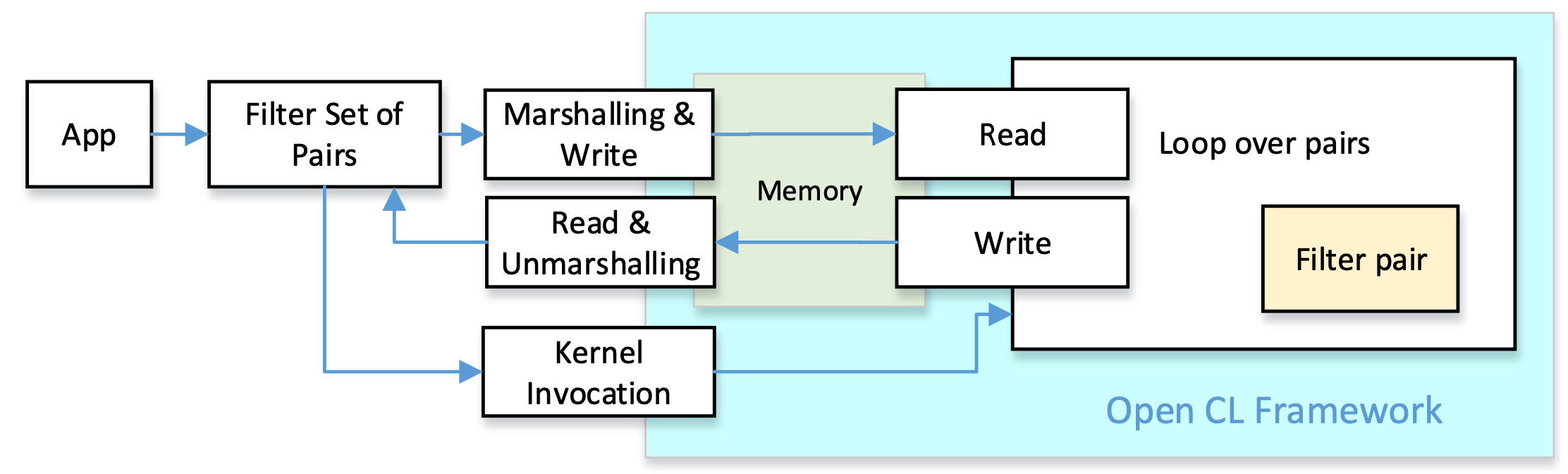
Our goal is to create pre-alignment filters that can be integrated into genomic datacenters equipped with FPGA coprocessors. We selected OpenCL as a widely available framework for the implementation of FPGA accelerators. All pre-alignment filters have the same goal: compare the pattern with a text, and obtain a distance estimation. This can be implemented in C/C++ by functions with the same number of parameters and the same return values. We can take profit of these similarities and implement a common set of functions for all filters both, at the host side and the accelerator side. The figure depicts the logic structure of the approach:
The algorithmic analysis to translate the existing proposals from HDL to C/C++ based HLS has the objective to explore method simplifications and optimization opportunities to produce more understandable algorithms with higher performance and lower resource usage.
