Next-Generation Sequencing (NGS) technologies have revolutionized many aspects of biology and personalized medicine. NGS systems significantly increase the throughput of DNA sequencing, while drastically reducing their cost.
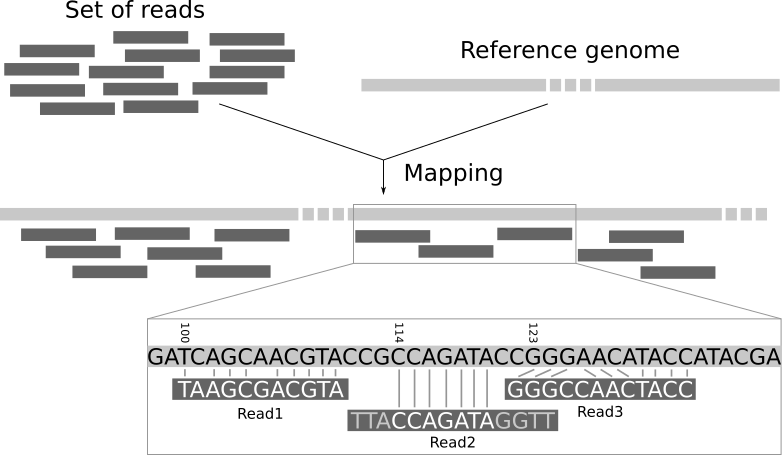
The data generated by these sequencing machines is organized in millions of small fragments called reads. In most sequence data analysis pipelines a main step is to determine the location of each read in the reference genome.
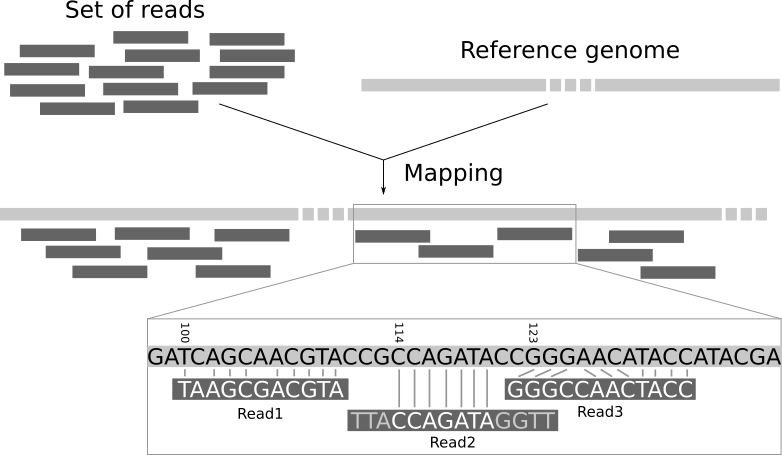
This problem is known as read-mapping or read-alignment. To solve it, we need to use specific software tools called mappers. In this context, the use of the recently proposed wavefront algorithm [2] allows the faster alignment of longer reads with a better efficiency due to its lower complexity. This is very relevant when considering third generation sequencing systems.
FPGAs are semiconductor devices that allow the actual functional definition of the devices in runtime with the use of their reconfiguration capabilities. These devices are usually personalized to accelerate the processing of some specific workloads with a very low energetic cost.
In the context of DRAC project, we present the first FPGA-based implementation of WFA algorithm. In a hardware/software co-designed scheme, the FPGA accelerator computes the alignment of pairs of sequences and generates the results in a compacted form that eases CPU-FPGA communication. Then, the CPU threads unpack the compacted forms to achieve the final results in parallel. The FPGA accelerator design is composed of multiple aligners that collaboratively compute the sequence alignments. The proposed design of the aligners allows a configurable maximum read length and error score between the reads. Thus, they can be adapted to the characteristics of the reads generated by different sequencing machines and technologies.
We evaluated the proposed WFA accelerator with different designs for typical read lengths and error score values on a high performance system with a POWER9 CPU and 2 FPGAs. Compared to the reference WFA CPU-only implementation, the FPGA accelerator achieves speedups of 4.5× to 8.8× with 1 FPGA, and of 8.2× to 13.5× with 2 FPGAs, while reducing the energy-to-solution by 6.1× to 9.7× with 1 FPGA, and by 11.4× to 14.6× with 2 FPGAs
===
1: Joachim Wolff, Bérénice Batut, Helena Rasche, 2021 Mapping (Galaxy Training Materials). /training-material/topics/sequence-analysis/tutorials/mapping/tutorial.html Online; accessed Tue Jun 01 2021
2: Marco-Sola, S., Moure, J. C., Moreto, M., & Espinosa, A. (2021). Fast gap-affine pairwise alignment using the wavefront algorithm. Bioinformatics, 37(4), 456-463.
3: A. Hagui et al., An FPGA Accelerator of the Wavefront Algorithm for Genomics Pairwise Alignment. FPL 2021. International Conference on Field-Programmable Logic and Applications. August 30th to September 3rd 2021. Technische Universität Dresden, Germany.
