Sequence alignment remains a fundamental problem with practical applications ranging from pattern recognition to computational biology. Traditional algorithms based on dynamic programming are hard to parallelize, require significant amounts of memory, and fail to scale for large inputs.
In the frame of the DRAC project, we have developed a GPU (graphics processing unit)-accelerated tool to compute the exact edit-distance sequence alignment based on the wavefront alignment algorithm (WFA) [1] . This approach exploits the similarities between the input sequences to accelerate the alignment process while requiring less memory than other algorithms.
The proposal presented [2] takes full advantage of the massive parallel capabilities of modern GPUs to accelerate the alignment process. In addition, we propose a succinct representation of the alignment data that successfully reduces the overall amount of memory required, allowing the exploitation of the fast shared memory of a GPU.
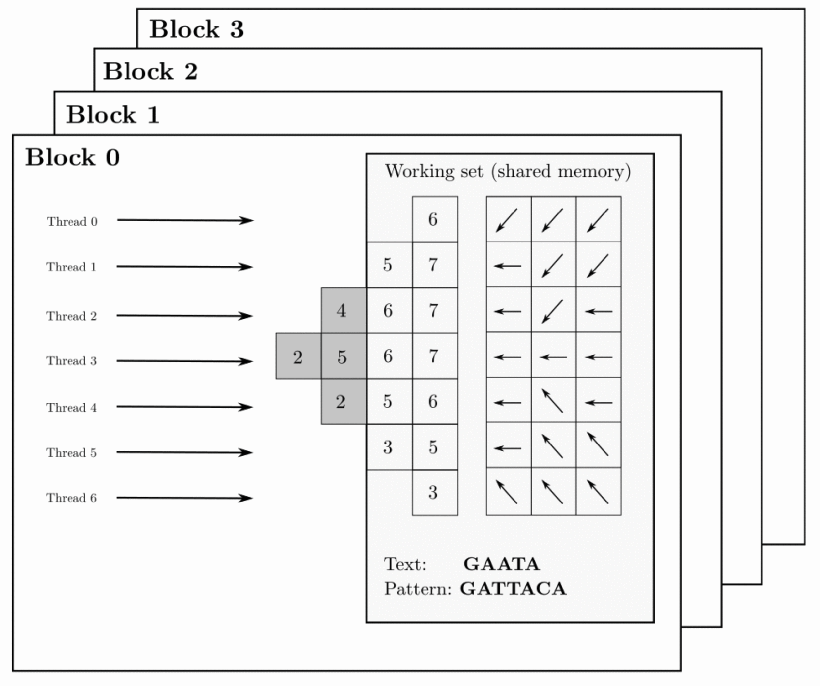
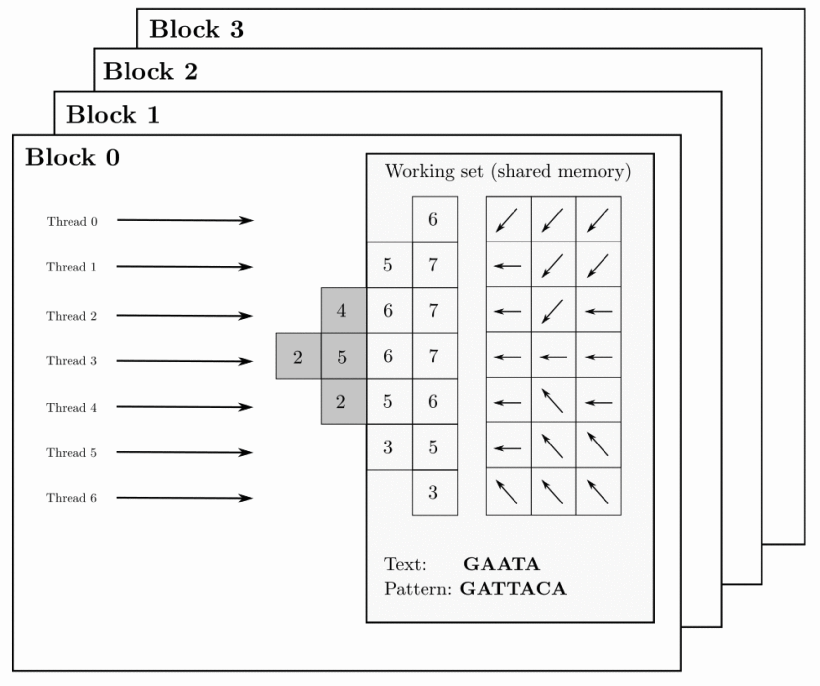
Nowadays, analysing large-scale workloads requires aligning millions of relatively large sequences to a given reference genome in a very short time. It is necessary to find a fine-grained strategy that computes each alignment using a thread block. This way, all threads within the block cooperatively work to compute one alignment problem. This new approach reduces the he consumption of shared memory and registers, allowing the storage of the wavefront structures in shared memory for several thread blocks, which can operate concurrently as shown in figure:
Our results show that our GPU implementation outperforms by 3-9× the baseline edit-distance WFA implementation running on a 20 core machine. As a result, eWFA-GPU is up to 265 times faster than state-of-the-art CPU implementation, and up to 56 times faster than state-of-the-art GPU implementations.
[1] Marco-Sola, S., Moure, J. C., Moreto, M., & Espinosa, A. (2021). Fast gap-affine pairwise alignment using the wavefront algorithm. Bioinformatics, 37(4), 456-463.
[2] Aguado-Puig, Q., Marco-Sola, S., Moure, J., Castells-Rufas, D., Alvarez, L., Espinosa, A., & Moreto, M. (2022). Accelerating Edit-Distance Sequence Alignment on GPU using the Wavefront Algorithm. IEEE access. 10, 63782-63796.
